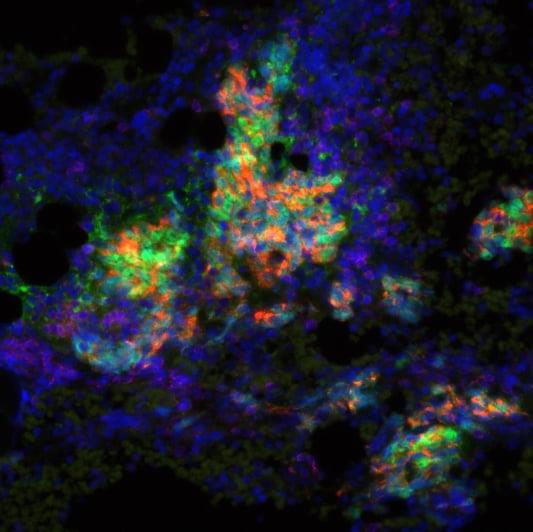
Exploiting new cancer vulnerabilities using chemical biology
Professor Edward Tate (Chemistry, Imperial College London), Professor Julian Downward (Formerly at The ICR), Dr Florence Raynaud (Cancer Therapeutics division, The ICR), Professor Holger W. Auner (Immunology and Inflammation, Imperial College London).
Predicting the immune response to new treatment using T-cell activity trackers
Dr Masahiro Ono (Life Science, Imperial College London), Professor Alan Melcher (Radiotherapy and Imaging, The ICR), Professor Kevin Harrington (Radiotherapy and Imaging, The ICR).
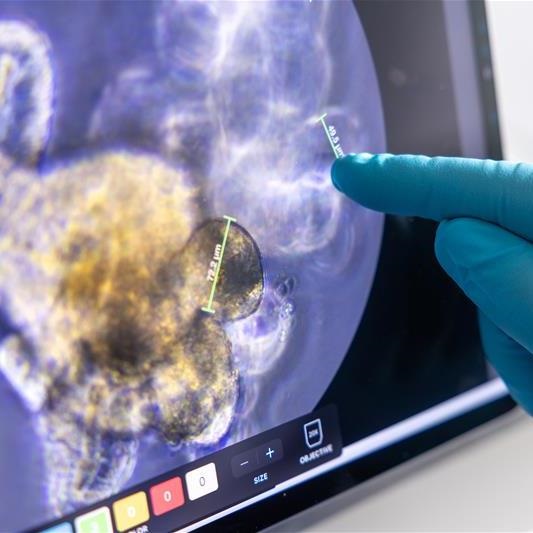
This project aimed to target cancer-reactive regulatory T cells (Tregs) to enhance precision immunotherapy. By combining the Tocky technology, which tracks T-cell activity over time, with expertise in oncolytic viruses and immune checkpoint blockade therapies, they explored new combination treatments for melanoma and refined targeting strategies (Bozhanova et al. J Immunother Cancer, 2022). Their findings highlight how T-cell signaling dynamics influence immunotherapy responses, providing insights to guide drug selection and improve future clinical trials. This led to another Centre-supported work showing that combination of the Maraba virus, which can kill cancer cells, and immune checkpoint inhibitors was able to overcome treatment resistance in difficult cases of advanced melanoma (Armstrong et al. J Immunother Cancer, 2024). Another study using Tocky highlighted the potential benefits of combining NKG2A blockers, PD-L1 inhibitors, ATR inhibitors, and radiotherapy (Patin et al. Nat. Commun, 2024), suggesting it as a promising combination strategies for treating head and neck squamous cell carcinoma.